R Speed with Ease
A Need for Speed
R often receives (unfair) criticism of being slow. In some cases this is true but often this is simply due to how the code is written and which packages are being leveraged. The R language is fast when vectorized functions are used (e.g. mean()), which avoids coding for loops in R. However, sometimes a problem is simply easier to code and understand when written as a loop; this is especially the case when the loops are nested. Other problems, such as running Markov chains, cannot be easily performed with vectorized functions. Or perhaps you have a high standard for highly performant code and want to take the reigns.
This post is a kind reminder to myself that, in many scenarios, it is easy to implement C++ code via the {Rcpp} package for extra performance or to simply allow a for loop to run free and without judgement. To get started (on Windows) is straight forward and only requires the same tools that R needs when installing packages from source. The focus here will be on the inline coding capability of {Rcpp} using cppFunction(), which can be great for simple tasks.
Example
The following example may not have great practical utility, nor be it the most optimized, but it does a good job showcasing the similarities between raw R and code written with Rcpp.
Imagine for the moment you are a data analyst who has been provided a vector of data. You want to perform a selective operation based upon the values in this vector. The values above zero will undergo a square root, whereas those zero and below will be raised to the power of two and then square rooted. As a piece-wise function in math jargon, this looks like:
\[ f(x) = \begin{cases} \sqrt{x} & : x > 0 \\ \sqrt{x^2} & : x \leq 0 \\ \end{cases} \]
However, you are uncertain as to what coding approach is most efficient and want to test out some options. Before we start defining the functions we must first load some packages and set the seed…
set.seed(9054)
library(Rcpp)
library(microbenchmark)
library(ggplot2)R vectorized
Typically, to solve this problem in R you would dip into vectorized solutions, perhaps using indexing, base R’s ifelse(), or fifelse() from {data.table}. The drawback to the latter instances in this particular use-case are they may produce warnings, which we will suppress later.
# Indexing
r_vec <- function(x) {
out <- vector('double', length(x))
idx <- x > 0
out[idx] <- sqrt(x[idx])
out[!idx] <- sqrt(x[!idx]^2)
out
}
# Base R ifelse
r_ifelse <- function(x) {
ifelse(x > 0, sqrt(x), sqrt(x^2))
}
# {data.table} equivalent of ifelse but faster
r_fifelse <- function(x) {
data.table::fifelse(x > 0, sqrt(x), sqrt(x^2))
}R loops
Perhaps, your brain works better using for loops. In base R this can be achieved quite simply by allocating a new vector and injecting the modified values.
# Slow R loop
r_loop <- function(x) {
out <- vector('double', length(x))
for(i in seq_along(x)) {
if(x[i] > 0) {
out[i] <- sqrt(x[i])
} else {
out[i] <- sqrt(x[i]^2)
}
}
return(out)
}Rcpp
Now we leverage {Rcpp} via cppFunction() to return a function named rcpp_loop. If you squint, you will see this looks very similar to the for loop from base R. The number of lines of code is also very similar. There are however a couple main differences of note:
- Indexing starts at 0, not 1
- Data requires a declared type (e.g.
NumericVector) - Semicolons (;) must be used as a line terminator
# Using C++ via Rcpp (function name: rcpp_loop)
Rcpp::cppFunction("
NumericVector rcpp_loop(NumericVector x) {
int n = x.size(); // Size of input vector
NumericVector out(n); // Create blank vector of equal size
for(int i = 0; i < n; i++) {
if(x[i] > 0) {
out[i] = sqrt(x[i]);
} else {
out[i] = sqrt(pow(x[i], 2.0));
}
}
return out;
}")Comparison
To compare the various functions, we generate a vector of 100,000 numbers from -500 to 500. To ensure all the functions return the same values, we pass this vector into each function and compare the results. For speed comparisons, we run microbenchmark 250 times for each function.
# Test data
my_vec <- runif(1e5, -500, 500)
# Check equality (returns content if all equal)
check_eq <- Reduce(\(x, y) if(all.equal(x, y)) x else FALSE,
x = list(rcpp_loop(my_vec),
r_vec(my_vec),
suppressWarnings(r_ifelse(my_vec)),
suppressWarnings(r_fifelse(my_vec)),
r_loop(my_vec))
)
# Benchmark
cpp_benchmark <- microbenchmark(rcpp = rcpp_loop(my_vec),
r_ifelse = suppressWarnings(r_ifelse(my_vec)),
r_fifelse = suppressWarnings(r_fifelse(my_vec)),
r_vec = r_vec(my_vec),
r_loop = r_loop(my_vec),
times = 250)
# Output results
print(cpp_benchmark, order = 'median')## Unit: microseconds
## expr min lq mean median uq max neval
## rcpp 421.0 445.3 541.5432 474.65 525.5 4442.3 250
## r_fifelse 1712.2 1867.1 2265.2816 1974.15 2152.8 7827.6 250
## r_vec 2079.3 2268.6 3243.7660 2617.30 2919.8 55034.8 250
## r_ifelse 2904.2 3174.9 4144.4700 3744.00 4076.6 9514.2 250
## r_loop 7694.0 7819.9 7956.5692 7893.70 7981.9 10615.7 250When sorted by the median run time across replications for each function we get a range from ~500 microseconds to ~8000 microseconds. Although the table is nice, it is easier to see the differences between replications and each function when presented in a plot.
boxplot(cpp_benchmark, log = TRUE, outline = TRUE)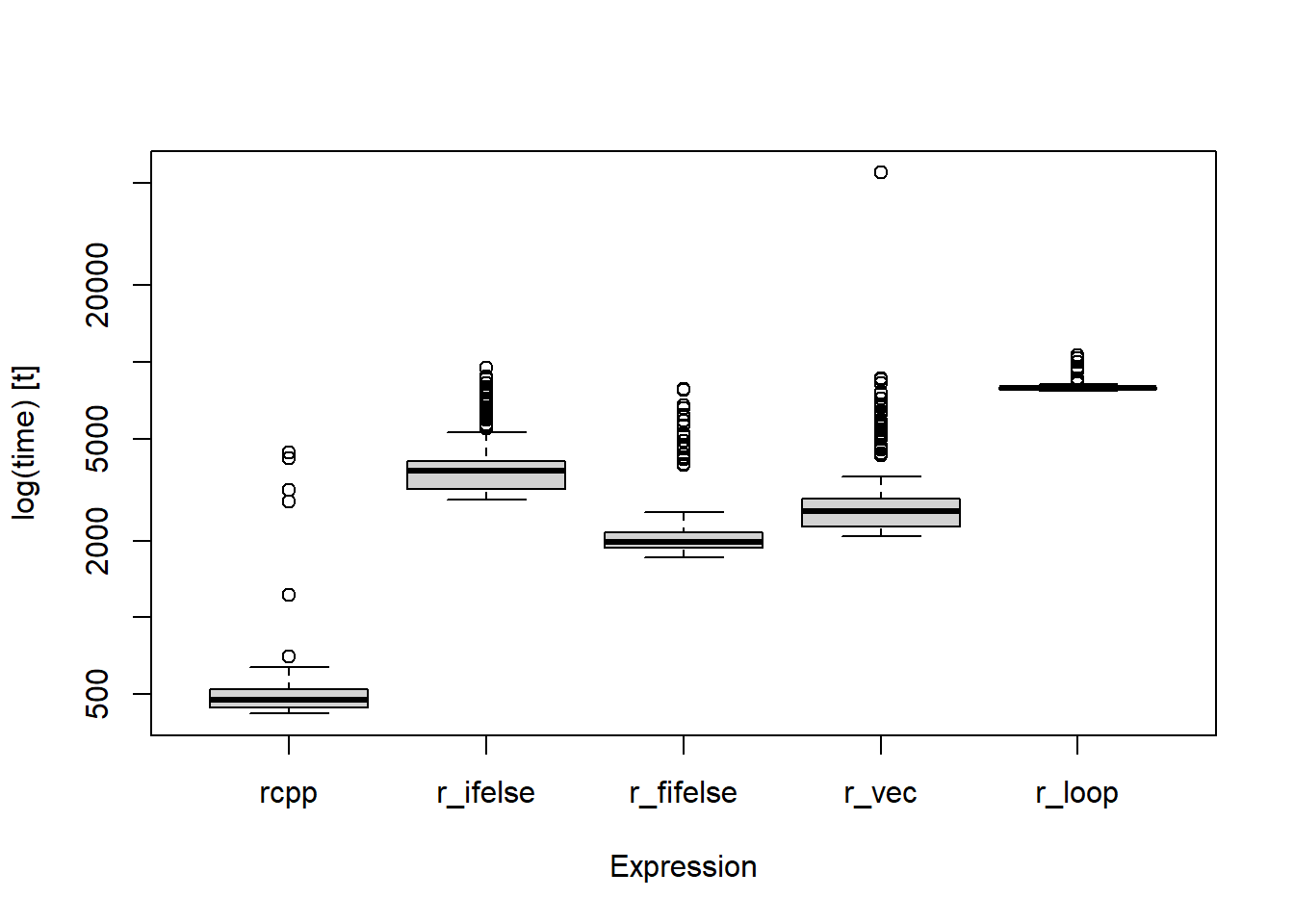
Figure 1: Comparison of different functions and their run times (logged microseconds) across 250 replications.
The {Rcpp} coded for loop was the fastest, the vectorized R function and datatable::fifelse() performed similarly, and the base R loop performed the worst overall (on my system anyway!). Using {Rcpp} can definitely make code run quickly, perhaps almost as quick as this dog…

End note
{Rcpp} provides an avenue to improve performance without having to leave the R ecosystem completely. The package comes with a number of rich features that make coding C++ code within R easy for someone who is primarily an R user, although it is not without its challenges. This post really only scratches the surface of {Rcpp} capabilities but often this is all one will need to get the speed boost they are looking for. There are plenty of resources available online that go into deeper details, a selection of which is provided below: