{relay}: an R Package for workflows
If I was being presumptuous, I would assume that the R programming language is primarily used for scripting tasks, especially in the field of Public Health. The R language is definitely capable of more complex tasks but, in terms of day-to-day use, creating scripts is most commonplace. Part of this is because R excels in this area, making it very easy for data analysts and epidemiologists to create reports quickly without the requirement to deeply understand software engineering principles and concepts. This is a good thing, making R very accessible.
There comes a point when you write an R script that is incredibly useful; this popularity soon demands more attention on not just what the scripts is doing but how it is run. Essentially, the code is being formalized and moving from development to production. Often there are relatively simple actions that can make this a less painful experience. At this point, being aware of some common software engineering principles is often necessary. Thankfully, this is a rather saturated area of development, so there are an abundance of tools available.
Workflow managment
As with most software, workflow management tools vary in complexity and purpose. Apache Airflow provides an open source option and is common among the Python community. The R package {targets} provides a similar solution to its respective community. There are also seemingly endless options specific for code tracing and logging; just a sample of these for R include: {loggit}, {futile.logger}, {tidylog}, and {lumberjack}. When I required this functionality, instead of using the existing solutions, I decided to create something new 🤷♂️.
What is {relay}?
{relay} is an R package for tracking workflows and their content. The original purpose of {relay} was to provide an easy method for scheduled RMarkdown files to be tied together and tracked. It grew to have a more general purpose with workflow organization and logging features that strikes a compromise in flexibility and structure. Considering the options already available, the existence of {relay} may seem unnecessary to those familiar with other frameworks. Creating {relay} made sense for our workflows given the team’s coding experience and the desire to continue to code as flexibly as possible. Getting started with {relay} should be as quick as basic logging but provide a more formal structure seen in more robust workflow management tools like {targets}. As such, {relay} may be a suitable alternative or middle ground for specific use-cases or perhaps used in conjunction with other workflow management and logging tools.
As the name suggests, {relay} invokes the image of a relay race. In order to track a particular process in an R script, a baton is created. After the particular step is complete, the baton is passed off and subsequently grabbed by any remaining steps in the process. As many batons as is sensible can be created to properly monitor scripts of interest.
The batons are S3 class objects with an associated YAML file which contain metadata (internal details from system), logbook, and content (workflow details from user). Since {relay} uses a YAML file and not just R objects, any software that can parse YAML files could use this information created from the R workflow. One can then conceive of workflows that may start in R and then inspected by and continued in python.
For additional information, take a look at the project README on GitHub.
{relay} in action
A workflow with dogs
To showcase {relay}, we will use it to monitor a very important workflow: extracting, parsing and reviewing dog names. We will be using the {rvest} package and a web-site to scrape a collection of dog names separated by gender and sorted alphabetically. For the sake of time, I will not explain the usage of {rvest} and instead focus on showcasing how {relay} monitors the workflow. If you are interested, check out the {rvest} package website to learn more.

First we load {relay} and other required packages, installing as necessary, and set our URL for dog names to a variable for safe keeping.
remotes::install_github('al-obrien/relay') # Run once
library(relay)
library(rvest)
library(magrittr)
# Base URL for dog names
base_url <- 'https://www.mera-petfood.com/en/dog/dogs-companion/'Start the relay!

Create the baton to track the workflow, by default it will create this baton in a temporary folder. Typically though, it would be preferable to assign to a more convenient location.
dog_baton <- create_baton()What does the baton contain? Well, let’s summarize some default details of our baton.
summary(dog_baton)##
## Metadata of baton...
## *id: 20221117124354-llCY1450
## *referee: TRACE
## *relay_type: COMPETITION
## *relay_start: 2022-11-17 12-43-54
## *relay_finish: NA
## *all_grabs:
## *all_passes:
## *pass_complete: FALSE
## *passes_completed: 0
## *location:
## C:\...\RtmpKanqll/_baton-20221117124354-llCY1450.yml
## *dropped: FALSE
##
## Contents of baton...
## *No contents.A unique ID is provided to the baton and a time-stamp of when the baton was created (i.e. the relay start time). Details on other metadata available are found here: help(create_baton). Currently, no content is in the bundle; however, as the workflow progresses, we may find some content should be captured in a structured way. We will add content later once we get going. In addition to baton content, we can also do some basic logging.
Using the log-book
The relay has begun and we want to log a message for our future selves to review. This is achieved by accessing the log-book. By setting the referee we can control what threshold of content will enter the baton’s logbook.
# Set threshold for the baton to capture messages or higher priority
set_referee(dog_baton, 'MESSAGE')# Write the important message (default is 'MESSAGE')
write_logbook(dog_baton, 'Starting my dog workflow!')
# Will be ignored
write_logbook(dog_baton, 'Checking a trace message', msg_type = 'TRACE')Let’s quickly review what is in the baton’s log-book!
read_logbook(dog_baton)## [1] "Pass [0] 2022-11-17 12:43:54 [MESSAGE] Starting my dog workflow!"The output probably looks like a familiar format, with the time-stamp, message type, and content listed. {relay} also includes the ‘pass’ under which the message was written. In this instance, the baton was just made, no pass was made yet, so it occurred during Pass [0].
Pass the baton
Now, let’s fetch the dog names (a-z) from our data source. Once completed, we will write another log message and then pass the baton. A baton can be passed and grabbed as often as we like; however, this action typically delineates essential components of the code. We can think of this like checkpoints in the workflow, where one operation is ‘passing off’ to the subsequent steps. If a checkpoint fails, we will know because that pass will be incomplete. In our example, we ‘encapsulate’ the web scraping step by passing the baton at the end.
# Read a-z pages of dog names
dog_page <- paste0('dog-names-', letters)
dog_names <- lapply(dog_page, function(x) read_html(paste0(base_url, x)))# Pass the baton after the process has completed
write_logbook(dog_baton, paste0('Dog pages (n: ', length(dog_names), ') a-z fetched.'))
dog_baton <- pass_baton(dog_baton)Let’s take a quick peek at our metadata again. We will see that is has been passed once successfully.
relay::read_metadata(dog_baton, subset = c('relay_start', 'all_passes', 'pass_complete'))## $relay_start
## [1] "2022-11-17 12-43-54"
##
## $all_passes
## [1] "2022-11-17 12-44-36"
##
## $pass_complete
## [1] TRUEGrab the baton
We now want to continue working with our freshly fetched (pun intended) dog names. To continue using {relay}, we grab the previously passed baton.
# The next 'player' in the relay grabs the baton previously passed
dog_baton <- grab_baton(dog_baton)Using a helper function for some basic parsing, we create our dog names data-set.
# Helper function to parse the page content
extract_both_gender <- function(names) {
tmp_list <- html_elements(names, '.list-eight-col')
tmp_list <- lapply(tmp_list, function(x) html_text(html_elements(x, 'li')))
return(data.frame('gender' = c(rep('male', length(tmp_list[[1]])),
rep('female', length(tmp_list[[2]]))),
'name' = c(tmp_list[[1]], tmp_list[[2]])))
}
# Bind all pages together to one dataset and create new column
dog_names_df <- do.call(rbind, lapply(dog_names, extract_both_gender))
dog_names_df$starts_with <- substr(dog_names_df$name, 1, 1)
write_logbook(dog_baton, paste0('Dog name dataset created (nrows: ', nrow(dog_names), ').'))With our dog name data in hand, we can now do some basic analysis…
# Count dog names by gender
n_gender <- tapply(dog_names_df$name, dog_names_df$gender, NROW)
dog_table <- knitr::kable(n_gender, col.names = 'Count (n)', caption = 'Number of dog names by gender.') %>%
kableExtra::scroll_box(box_css = "",
extra_css = 'overflow-x: auto; width: 50%')
dog_table| Count (n) | |
|---|---|
| female | 3991 |
| male | 5732 |
# Count by starting character
N_st_char <- tapply(dog_names_df$gender, dog_names_df$starts_with, NROW)
N_st_char_sorted <- sort(N_st_char, decreasing = TRUE)
barplot(N_st_char_sorted,
cex.names = .7,
xlab = 'Dog name starts with...',
ylab = 'Count (n)',
col = 'lightblue',
border = FALSE)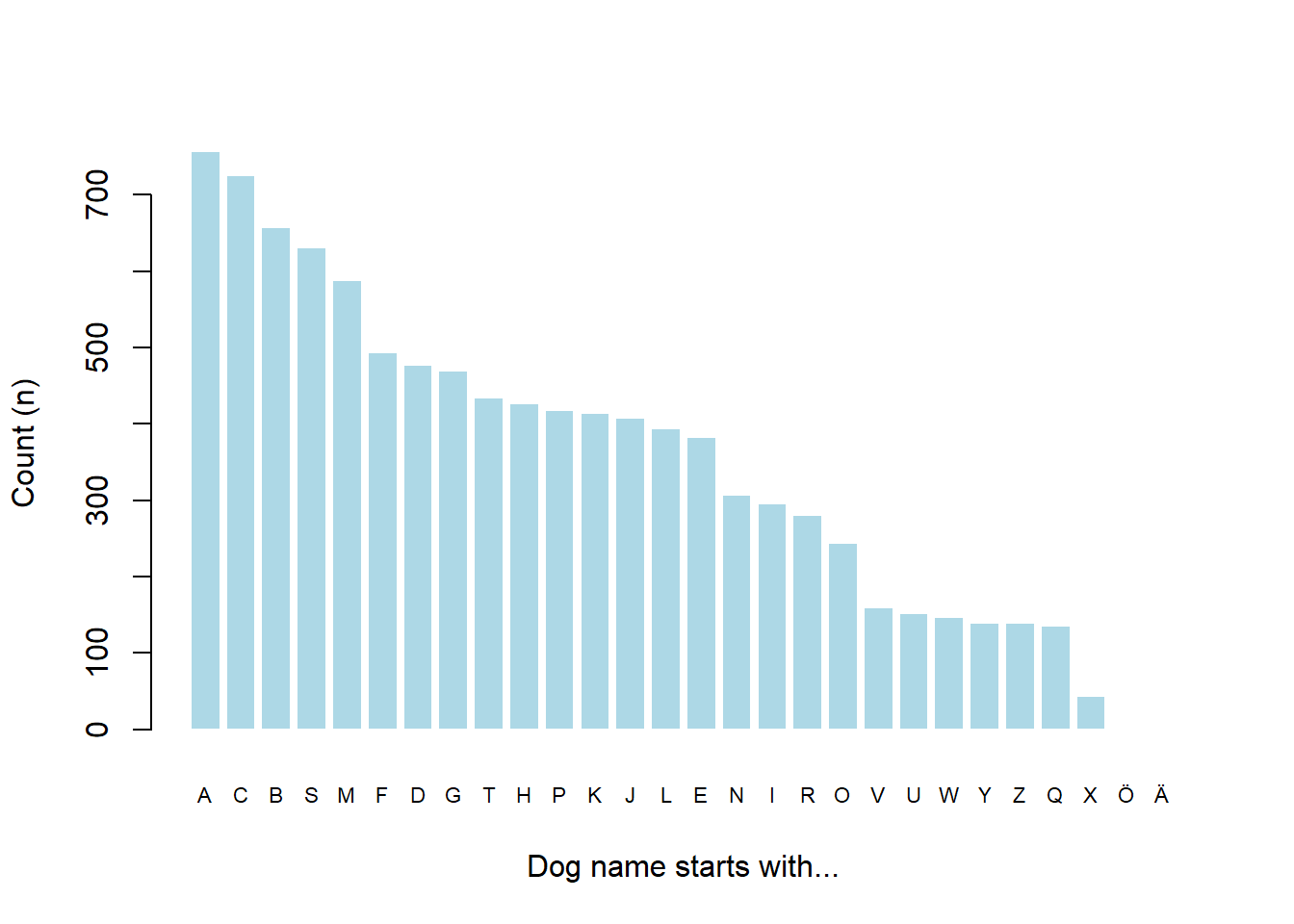
Figure 1: Number of dog names starting with a particular letter.
Excellent! Most dog names start with the letter ‘A’ and there are quite a few more male dog names when compared to female (in this particular list anyway). We have also logged the efforts to discover this along the way.
Bundling up with {relay}

Our workflow, albeit basic, now has several objects. We may start to realize that having many inputs, R objects, and outputs is becoming unwieldy. {relay} provides several helper-functions to assist in organizing more complex workflows. The main operation is to create a bundle of content. There are some benefits in bundling data, code, and metadata in a single location. For one, it allows the entire set to be zipped and shared as a (mainly) reproducible environment. If the workflow is repeated on a schedule, then each instance can be isolated as independent sets.
Even if we only want to run our dog name workflow once, let’s still place all relevant output files in a bundle. First, we use the existing baton (dog_baton) as a parameter to create_bundle() and save it under a temporary directory. We use the tree parameter to assign a nested ‘outputs’ folder. We then relocate the baton to inside the bundle (which follows a similar naming convention) using a dedicated helper function. Various other helper and utility functions are available (often acting as thin wrappers) to make moving files a bit easier (e.g., quick_copy(), copy_files(), copy_dir(), load_batch()).
# Assign a temporary spot
temp_wd <- tempdir()
# Make a bundle to place baton and other details
bundled_baton <- create_bundle(dog_baton, dir = temp_wd, tree = 'outputs')
dog_baton <- relay:::relocate_baton(dog_baton, bundled_baton)We could have also created a bundle right from the start by setting bundled = TRUE!
bundled_baton <- create_baton(bundled = TRUE, bundle_params = list(dir = temp_wd, tree = NULL))Since we adore all things about dogs, we feel attached to this data and must save it persistently. We do so by writing dog_names_df to a CSV file and the raw extract dog_names as a RDS file within the bundle.
# Write data.frame to the bundle under outputs
out_path <- file.path(bundled_baton, 'outputs', 'dog_name.csv')
write.csv(dog_names_df, out_path, row.names = FALSE)
# Save raw extract too
saveRDS(dog_names, file.path(bundled_baton, 'outputs', 'dog_names_raw.rds'))
# Check the bundle structure
fs::dir_tree(bundled_baton)Add baton content
With our basic workflow now complete, we perform one final pass but this time we add some content to the baton. In this example I use vapply() but others may prefer the purrr family. We add vectors that confirm the data existence and creation times. We also see the final baton was stamped as ‘complete’ and performed two passes. This can be useful for scripts dependent on this process; they can read the metadata in the baton and confirm it was completed successfully and that all the necessary data is available (and ‘fresh’). Since we are using a YAML file for the baton, it is easy for another process to parse the content fields (as compared to the log-book).
data_names <- c('dog_name.csv', 'dog_names_raw.rds')
# Create some information about the data to include in the bundle
existence <- vapply(data_names,
function(x) file.exists(paste0(bundled_baton, '/outputs/', x)),
logical(1),
USE.NAMES = FALSE)
creation_times <- vapply(data_names,
function(x) as.character(file.info(paste0(bundled_baton, '/outputs/', x))$ctime),
character(1),
USE.NAMES = FALSE)
# Define content of baton to save to YAML in addition to metadata (nested lists)
baton_content <- list(datasets = list(names = data_names,
existence = existence,
creation_times = creation_times))
# Perform final pass with the custom content added
dog_baton <- pass_baton(dog_baton, content = baton_content)
summary(dog_baton)##
## Metadata of baton...
## *id: 20221117124354-llCY1450
## *referee: MESSAGE
## *relay_type: COMPETITION
## *relay_start: 2022-11-17 12-43-54
## *relay_finish: 2022-11-17 12-44-37
## *all_grabs: 2022-11-17 12-44-36
## *all_passes: 2022-11-17 12-44-36
## *all_passes: 2022-11-17 12-44-37
## *pass_complete: TRUE
## *passes_completed: 2
## *location:
## C:\...\RtmpKanqll\_bundle-20221117124354-llCY1450/_baton-20221117124354-llCY1450.yml
## *dropped: FALSE
##
## Contents of baton...
## *datasets: c("dog_name.csv", "dog_names_raw.rds")
## *datasets: c(TRUE, TRUE)
## *datasets: c("2022-11-17 12:44:16", "2022-11-17 12:44:17")Wrap up…
We have used {relay} to provide various logging details and to help organize aspects of our workflow to discover facts about dog names. Hopefully this, rather contrived, example provides a good overview that encourages you to give {relay} a try in your next project, be it about dogs or not (perhaps cats…)!
The code below shows an uninterrupted example of the dog name workflow discussed above.
#------------------#
# Libraries
#------------------#
library(rvest)
library(relay)
library(magrittr)
#------------------#
# Init baton & params
#------------------#
# Create baton for process and et threshold for the baton
dog_baton <- create_baton()
set_referee(dog_baton, 'MESSAGE')
write_logbook(dog_baton, 'Starting my dog workflow!')
# Helper function to extract dog names by gender
extract_both_gender <- function(names) {
tmp_list <- rvest::html_elements(names, '.list-eight-col')
tmp_list <- lapply(tmp_list, function(x) rvest::html_text(rvest::html_elements(x, 'li')))
return(data.frame('gender' = c(rep('male', length(tmp_list[[1]])),
rep('female', length(tmp_list[[2]]))),
'name' = c(tmp_list[[1]], tmp_list[[2]])))
}
# Base URL for dog names
base_url <- 'https://www.mera-petfood.com/en/dog/dogs-companion/'
#------------------#
# Web scrape
#------------------#
# Pull all dog names
dog_page <- paste0('dog-names-', letters)
dog_names <- lapply(dog_page, function(x) read_html(paste0(base_url, x)))
#------------------#
# Parse and create df
#------------------#
# Bind all pages together to one dataset and create new column
dog_names_df <- do.call(rbind, lapply(dog_names, extract_both_gender))
dog_names_df$starts_with <- substr(dog_names_df$name, 1, 1)
write_logbook(dog_baton, 'Dog name dataset created (nrows: ' (nrow(dog_names)), ').')
#------------------#
# Analyze
#------------------#
# Count dog names by gender
n_gender <- tapply(dog_names_df$name, dog_names_df$gender, NROW)
dog_table <- knitr::kable(n_gender, col.names = 'Count (n)', caption = 'Number of dog names by gender.')
# Count by starting character
N_st_char <- tapply(dog_names_df$gender, dog_names_df$starts_with, NROW)
N_st_char_sorted <- sort(N_st_char, decreasing = TRUE)
barplot(N_st_char_sorted,
cex.names = .7,
xlab = 'Dog name starts with...',
ylab = 'Count (n)',
col = 'lightblue',
border = FALSE)
dog_plot <- recordPlot()
#------------------#
# Outputs & finish
#------------------#
# Write data.frame to the bundle under outputs
out_path <- file.path(bundled_baton, 'outputs', 'dog_name.csv')
write.csv(dog_names_df, out_path, row.names = FALSE)
# Save raw extract too
saveRDS(dog_names, file.path(bundled_baton, 'outputs', 'dog_name_raw.rds'))
data_names <- c('dog_name.csv', 'dog_names_raw.rds')
# Create some information about the data to include in the bundle
existence <- vapply(data_names,
function(x) file.exists(paste0(bundled_baton, '/outputs/', x)),
logical(1),
USE.NAMES = FALSE)
creation_times <- vapply(data_names,
function(x) as.character(file.info(paste0(bundled_baton, '/outputs/', x))$ctime),
character(1),
USE.NAMES = FALSE)
# Define content of baton to save to YAML in addition to metadata (nested lists)
baton_content <- list(datasets = list(names = data_names,
existence = existence,
creation_times = creation_times))
# Perform final pass with the custom content added
dog_baton <- pass_baton(dog_baton, content = baton_content)